Wout Bittremieux, PhD
Assistant research professor

University of Antwerp
Campus Middelheim
Middelheimlaan 1
2020 Antwerpen
Belgium
- bittremieux
- 0000-0002-3105-1359
- wout DOT bittremieux AT uantwerpen DOT be
Prof. Dr. Wout Bittremieux is an assistant research professor in the Adrem Data Lab at the University of Antwerp. His research employs advanced computational techniques to solve fundamental biological questions, by developing algorithmic solutions and machine learning methods for the analysis of mass spectrometry-based proteomics and metabolomics data. He has developed several innovative tools to harness novel biological insights from hundreds of millions of mass spectra. His work is ideally positioned at the intersection of bioinformatics, machine learning, and mass spectrometry.
In 2020, Dr. Bittremieux received the prestigious Postdoctoral Career Development Award from the American Society for Mass Spectrometry, in 2021 he was named a "Rising Star in Proteomics and Metabolomics" by the Journal of Proteome Research for demonstrating 'incredible originality and promise for the future of proteomics and metabolomics,' in 2022 he won the Early Career Researcher Manuscript Competition by the Human Proteome Organization, in 2023 he received the Bioinformatics for Mass Spectrometry Award from the European Proteomics Association and the Metabolites Young Investigator Award, and in 2025 he received the Junior Research Parasite Award.
Dr. Bittremieux is an avid contributor to the computational mass spectrometry community. He has published widely in internationally recognized scientific journals, is an active member of the European Bioinformatics Community for Mass Spectrometry (EuBIC-MS), and leads the CompMS interest group of the International Society for Computational Biology (ISCB). Furthermore, he has contributed to the development of mass spectrometry data standards defined by the Proteomics Standards Initiative (PSI) and is an active developer of the Global Natural Products Social Molecular Networking (GNPS) platform, through which his tools reach tens of thousands of monthly users worldwide.
Publications
See also Google Scholar.
2026
-
Journal The 2025 Westlake Autumn Symposium for AI Proteomics and Virtual Cell.
Genomics, Proteomics & Bioinformatics, In Press (2026)
-
Journal Strategic priorities for transformative progress in advancing biology with proteomics and artificial intelligence.
Nature Methods, In Press (2026)
-
Journal Tracing the origins of molecular signals in food through integrative metabolomics and chemical databases.
npj Science of Food, In Press (2026)
-
Journal pmultiqc: An open-source, lightweight, and metadata-oriented QC reporting library for MS proteomics.
Molecular & Cellular Proteomics (2026)
-
Journal Scalable incremental clustering for tandem mass spectra in untargeted metabolomics.
Journal of Proteome Research (2026)
-
Journal An open-source platform for reference data-driven analysis of untargeted metabolomics.
Journal of the American Society for Mass Spectrometry (2026)
-
Journal Improved open modification searching via unified spectral search with predicted libraries and enhanced vector representations in ANN-SoLo.
Journal of Proteome Research (2026)
-
Preprint Scalable mass-spectrometry-based molecular phylogeny with TreeMS2.
bioRxiv (2026)
-
Preprint Hardware software synergy in proteomics: Systematic evaluation across mass spectrometry platforms, search engines, and rescoring methods.
ChemRxiv (2026)
-
Preprint An interactive web application for composing nearest-neighbor suspect spectral libraries.
ChemRxiv (2026)
-
Journal SquiDBase: A community resource of raw nanopore data from microbes.
NAR Genomics and Bioinformatics 8, lqaf213 (2026)
-
Journal Improvements to Casanovo, a deep learning de novo peptide sequencer.
Journal of Proteome Research 25, 755–764 (2026)
-
Preprint Positive-unlabeled learning for predicting small molecule MS2 Identifiability from MS1 context and acquisition parameters.
bioRxiv (2026)
2025
-
Journal Scalability of mass spectrometry-based metabolomics for natural extracts libraries exploration: Current status, challenges, and opportunities.
Natural Product Reports, Advance Article (2025)
-
Journal Self-supervised learning from small-molecule mass spectrometry data.
Nature Biotechnology (2025)
-
Journal Perspectives in computational mass spectrometry: Recent developments and key challenges.
Bioinformatics Advances 5, vbaf301 (2025)
-
Preprint ProteoBench: The community-curated platform for comparing proteomics data analysis workflows.
bioRxiv (2025)
-
Journal A resource to empirically establish drug exposure records directly from untargeted metabolomics data.
Nature Communications 16, 10600 (2025)
-
Journal mzPeak: Designing a scalable, interoperable, and future-ready mass spectrometry data format.
Journal of Proteome Research 24, 5329–5335 (2025)
-
Preprint Bridging complexity and accessibility in metabolomics with MetaboApps.
ChemRxiv (2025)
-
Journal Comparing the applicability of de facto population markers for spatiotemporal trend analysis in wastewater-based epidemiology.
Journal of Hazardous Materials 496, 139332 (2025)
-
Journal A 2025 perspective on the role of machine learning for biomarker discovery in clinical proteomics.
Expert Review of Proteomics 22, 363–374 (2025)
-
Preprint Temporal regulation of metabolic processes in the marine diazotroph Crocosphaera watsonii WH 8501.
bioRxiv (2025)
-
Journal A universal language for finding mass spectrometry data patterns.
Nature Methods 22, 1247–1254 (2025)
-
Journal SpectriPy: Enhancing cross-language mass spectrometry data analysis with R and Python.
Journal of Open Source Software 10, 8070 (2025)
-
Preprint Foundation model for mass spectrometry proteomics.
arXiv:2505.10848 (2025)
-
Preprint MetaboT: An LLM-based multi-agent framework for interactive analysis of mass spectrometry metabolomics knowledge.
Research Square (2025)
-
Journal Open-source and FAIR research software for proteomics.
Journal of Proteome Research 24, 2222–2234 (2025)
-
Journal MS-RT: A method for evaluating MS/MS clustering performance for metabolomics data.
Journal of Proteome Research 24, 1778–1790 (2025)
-
Journal pyOpenMS-viz: Streamlining mass spectrometry data visualization with pandas.
Journal of Proteome Research 24, 2152–2158 (2025)
-
Journal Discovery of metabolites prevails amid in-source fragmentation.
Nature Metabolism 7, 435–437 (2025)
-
Journal Molecular structure discovery for untargeted metabolomics using biotransformation rules and global molecular networking.
Analytical Chemistry 97, 3213–3219 (2025)
-
Journal Comparison of in vitro biotransformation of olive polyphenols between healthy young and elderly.
Metabolites 15, 26 (2025)
2024
-
Preprint Zero-shot retention time prediction for unseen post-translational modifications with molecular structure encodings.
bioRxiv (2024)
-
Proceeding MassSpecGym: A benchmark for the discovery and identification of molecules.
Proceedings of the 38th Conference on Neural Information Processing Systems (NeurIPS 2024) Datasets and Benchmarks Track (2024)
-
Journal Deep learning methods for
de novo peptide sequencing.Mass Spectrometry Reviews, early view (2024)
-
Journal The Proteomics Standards Initiative standardized formats for spectral libraries and fragment ion peak annotations: mzSpecLib and mzPAF.
Analytical Chemistry 96, 18491–18501 (2024)
-
Proceeding RapidOMS: FPGA-Based open modification spectral library searching with HD computing.
Proceedings of the 2024 IEEE Biomedical Circuits and Systems Conference - BioCAS 2024 (2024)
-
Journal Accounting for digestion enzyme bias in Casanovo.
Journal of Proteome Research 23, 4761–4769 (2024)
-
Journal Reusability report: Annotating metabolite mass spectra with domain-inspired chemical formula transformers.
Nature Machine Intelligence 6, 1296–1302 (2024)
-
Journal Machine learning strategies to tackle data challenges in mass spectrometry-based proteomics.
Journal of the American Society for Mass Spectrometry 35, 2143–2155 (2024)
-
Journal DRAM-based acceleration of open modification search for mass spectrometry-based proteomics.
IEEE Transactions on Computer-Aided Design of Integrated Circuits and Systems 43, 2592–2605 (2024)
-
Journal Semisupervised learning to boost hERG, Nav1.5, and Cav1.2 cardiac ion channel toxicity prediction by mining a large unlabeled small molecule data set.
Journal of Chemical Information and Modeling 64, 6410–6420 (2024)
-
Journal Communicating mass spectrometry quality information in mzQC with Python, R, and Java.
Journal of the American Society for Mass Spectrometry 35, 1875–1882 (2024)
-
Journal Sequence-to-sequence translation from mass spectra to peptides with a transformer model.
Nature Communications 15, 6427 (2024)
-
Journal Fragment ion intensity prediction improves the identification rate of non-tryptic peptides in timsTOF.
Nature Communications 15, 3956 (2024)
-
Journal Machine learning-based peptide-spectrum match rescoring opens up the immunopeptidome.
PROTEOMICS 24, 2300336 (2024)
-
Journal From data to discovery: The essential role of computational tools in proteomics.
PROTEOMICS 24, 2300081 (2024)
-
Journal Benchmarking of small molecule feature representations for hERG, Nav1.5, and Cav1.2 cardiotoxicity prediction.
Journal of Chemical Information and Modeling 64, 2515–2527 (2024)
-
Journal The underappreciated diversity of bile acid modifications.
Cell 187, 1801–1818 (2024)
-
Proceeding SpecHD: Hyperdimensional computing framework for FPGA-based mass spectrometry clustering.
Proceedings of the 2024 Design, Automation & Test in Europe Conference - DATE 2024 (2024)
-
Journal Open access repository-scale propagated nearest neighbor suspect spectral library for untargeted metabolomics.
Nature Communications 14, 8488 (2023)
"Top 25 Health Sciences Articles of 2023" by Nature Communications
2025 Junior Research Parasite Award
-
Preprint A cross-attention transformer encoder for paired sequence data.
bioRxiv (2023)
-
Journal The pitfalls of negative data bias for the T-cell epitope specificity challenge.
Nature Machine Intelligence 5, 1060–1062 (2023)
-
Journal Accelerating open modification spectral library searching on tensor core in high-dimensional space.
Bioinformatics 22, btad404 (2023)
-
Journal Interpretable deep learning to uncover the molecular binding patterns determining TCR–epitope interactions.
ImmunoInformatics 11, 100027 (2023)
-
Journal HyperSpec: Ultra-fast mass spectra clustering in hyperdimensional space.
Journal of Proteome Research 22, 1639–1648 (2023)
-
Journal Unified and standardized mass spectrometry data processing in Python using spectrum_utils.
Journal of Proteome Research 22, 625–631 (2023)
-
Journal Semisupervised machine learning for sensitive open modification spectral library searching.
Journal of Proteome Research 22, 585–593 (2023)
-
Journal The Proteomics Standards Initiative at twenty years: Current activities and future work.
Journal of Proteome Research 22, 287–301 (2023)
2023
2022
-
Journal Open modification searching of SARS-CoV-2-human protein interaction data reveals novel viral modification sites.
Molecular & Cellular Proteomics 21, 100425 (2022)
-
Journal Standardized multi-omics of Earth's microbiomes reveals microbial and metabolite diversity.
Nature Microbiology 7, 2128-2150 (2022)
-
Journal The critical role that spectral libraries play in capturing the metabolomics community knowledge.
Metabolomics 18, 94 (2022)
-
Journal Four layer multi-omics reveals molecular responses to aneuploidy in Leishmania.
PLOS Pathogens 18, e1010848 (2022)
-
Proceeding Massively parallel open modification spectral library searching with hyperdimensional computing.
Proceedings of the 31st International Conference on Parallel Architectures and Compilation Techniques - PACT 2022 (2022)
-
Journal Comparison of cosine, modified cosine, and neutral loss based spectral alignment for discovery of structurally related molecules.
Journal of the American Society for Mass Spectrometry 33, 1733–1744 (2022)
-
Journal Native metabolomics identifies the rivulariapeptolide family of protease inhibitors.
Nature Communications 13, 4619 (2022)
-
Proceeding De novo mass spectrometry peptide sequencing with a transformer model.
Proceedings of the 39th International Conference on Machine Learning - ICML 2022 (2022)
-
Journal Enhancing untargeted metabolomics using metadata-based source annotation.
Nature Biotechnology 40, 1774–1779 (2022)
-
Journal A learned embedding for efficient joint analysis of millions of mass spectra.
Nature Methods 19, 675–678 (2022)
"Early Career Researcher Manuscript Competition" winner by the Human Proteome Organization
-
Journal A neural network for large-scale clustering of peptide mass spectra.
Nature Methods 19, 658–659 (2022)
-
Journal A comprehensive evaluation of consensus spectrum generation methods in proteomics.
Journal of Proteome Research 21, 1566–1574 (2022)
-
Journal Proteomics Standards Initiatives ProForma 2.0: Unifying the encoding of Proteoforms and Peptidoforms.
Journal of Proteome Research 21, 1189–1195 (2022)
-
Journal Physicochemical properties determining drug detection in skin.
Clinical and Translational Science 15, 761–770 (2022)
2021
-
Journal ppx: Programmatic access to proteomics data repositories.
Journal of Proteome Research 20, 4621–4624 (2021)
-
Journal Current challenges for unseen-epitope TCR interaction prediction and a new perspective derived from image classification.
Briefings in Bioinformatics 22, bbaa318 (2021)
-
Journal Universal Spectrum Identifier for mass spectra.
Nature Methods 18, 768–770 (2021)
-
Journal Large-scale tandem mass spectrum clustering using fast nearest neighbor searching.
Rapid Communications in Mass Spectrometry 39, e9153 (2021)
-
Journal Current and future deep learning algorithms for MS/MS-based small molecule structure elucidation.
Rapid Communications in Mass Spectrometry 39, e9120 (2021)
-
Journal The European Bioinformatics Community for Mass Spectrometry (EuBIC-MS): An open community for bioinformatics training and research.
Rapid Communications in Mass Spectrometry 39, e9087 (2021)
-
Journal Sharing biological data: Why, when, and how.
FEBS Letters 595, 847–863 (2021)
-
Journal Open science resources for the mass spectrometry-based analysis of SARS-CoV-2.
Journal of Proteome Research 20, 1464–1475 (2021)
-
Journal Auto-deconvolution and molecular networking of gas chromatography–mass spectrometry data.
Nature Biotechnology 39, 169–173 (2021)
2020
-
Journal Proceedings of the EuBIC-MS 2020 Developers' Meeting.
EuPA Open Proteomics 24, 1–6 (2020)
-
Preprint Universal MS/MS visualization and retrieval with the Metabolomics Spectrum Resolver web service.
bioRxiv (2020)
-
Book Identification of epitope-specific T cells in T-cell receptor repertoires.
Bioinformatics for Cancer Immunotherapy 2120, 183–195 (2020)
-
Journal MESSAR: Automated recommendation of metabolite substructures from tandem mass spectra.
PLOS ONE 15, e0226770 (2020)
-
Journal spectrum_utils: A Python package for mass spectrometry data processing and visualization.
Analytical Chemistry 92, 659–661 (2020)
2019
-
Journal Detection of enriched T cell epitope specificity in full T cell receptor sequence repertoires.
Frontiers in Immunology 10, 2820 (2019)
-
Journal Mining the enriched subgraphs for specific vertices in a biological graph.
IEEE/ACM Transactions on Computational Biology and Bioinformatics 16, 1496–1507 (2019)
-
Journal 2018 YPIC Challenge: A case study in characterizing an unknown protein sample.
Journal of Proteome Research 18, 3936–3943 (2019)
-
Journal Extremely fast and accurate open modification spectral library searching of high-resolution mass spectra using feature hashing and graphics processing units.
Journal of Proteome Research 18, 3792–3799 (2019)
-
Journal Proceedings of the EuBIC Winter School 2019.
EuPA Open Proteomics 22–23, 4–7 (2019)
-
Journal Using expert driven machine learning to enhance dynamic metabolomics data analysis.
Metabolites 9, 54 (2019)
2018
-
Journal Fast open modification spectral library searching through approximate nearest neighbor indexing.
Journal of Proteome Research 17, 3463–3474 (2018)
"Rising Star in Proteomics and Metabolomics" awarded by the Journal of Proteome Research
-
Journal Proceedings of the EuBIC Developer's Meeting 2018.
Journal of Proteomics 187, 25–27 (2018)
-
Journal Grasping frequent subgraph mining for bioinformatics applications.
BioData Mining 11 (2018)
-
Journal Quality control in mass spectrometry-based proteomics.
Mass Spectrometry Reviews 37, 697–711 (2018)
-
Journal On the feasibility of mining CD8+ T-cell receptor patterns underlying immunogenic peptide recognition.
Immunogenetics 70, 159–168 (2018)
2017
-
Journal Proteomics Standards Initiative: Fifteen years of progress and future work.
Journal of Proteome Research 16, 4288–4298 (2017)
-
Journal A community proposal to integrate proteomics activities in ELIXIR [version 1; referees: 2 approved].
F1000Research 6, 875 (2017)
-
Journal The Human Proteome Organization–Proteomics Standards Initiative Quality Control working group: Making quality control more accessible for biological mass spectrometry.
Analytical Chemistry 89, 4474–4479 (2017)
-
Thesis Computational solutions for quality control of mass spectrometry-based proteomics.
Universiteit Antwerpen (2017)
-
Journal Computational quality control tools for mass spectrometry proteomics.
PROTEOMICS 17, 1600159 (2017)
2016
-
Journal Designing biomedical proteomics experiments: State-of-the-art and future perspectives.
Expert Review of Proteomics 13, 495–511 (2016)
-
Journal Unsupervised quality assessment of mass spectrometry proteomics experiments by multivariate quality control metrics.
Journal of Proteome Research 15, 1300–1307 (2016)
2015
-
Proceeding Discovery of significantly enriched subgraphs associated with selected vertices in a single graph.
Proceedings of the 14th International Workshop on Data Mining in Bioinformatics - BIOKDD '15 (2015)
-
Journal iMonDB: Mass spectrometry quality control through instrument monitoring.
Journal of Proteome Research 14, 2360–2366 (2015)
-
Journal A primer to frequent itemset mining for bioinformatics.
Briefings in Bioinformatics 16, 216–231 (2015)
2014
-
Journal Efficient reduction of candidate matches in peptide spectrum library searching using the top k most intense peaks.
Journal of Proteome Research 13, 4175–4183 (2014)
-
Journal qcML: An exchange format for quality control metrics from mass spectrometry experiments.
Molecular & Cellular Proteomics 13, 1905–1913 (2014)
-
Journal jqcML: An open-source Java API for mass spectrometry quality control data in the qcML format.
Journal of Proteome Research 13, 3484–3487 (2014)
-
Journal Machine learning applications in proteomics research: How the past can boost the future.
PROTEOMICS 14, 353–366 (2014)
Presentations
2026
-
Invited Learning the rules of molecular behavior from open mass spectrometry repositories.
Biophysical Society, San Francisco, CA, USA (February 24, 2026)
-
Invited Cracking the mass spectrometry code: Artificial intelligence for de novo peptide sequencing.
Festival of Genomics & Biodata, London, United Kingdom (January 29, 2026)
2025
-
Invited Verborgen moleculaire werelden ontrafeld: AI om onze voeding en gezondheid in kaart te brengen.
Wetenschapscafé, Antwerp, Belgium (December 17, 2025)
-
Invited Understanding the molecular phenotype with AI for proteomics and metabolomics.
AntwerpHealth.AI, Antwerp, Belgium (December 9, 2025)
-
Invited AI-powered computational mass spectrometry for molecular discovery.
Microbiotec 2025, Ponta Delgada, Portugal (December 6, 2025)
-
Invited AI om het moleculaire universum te ontrafelen.
Volkssterrenwacht Urania, Hove, Belgium (November 25, 2025)
-
Invited From open mass spectrometry data to molecular discovery.
Open Access Week Norway (October 23, 2025)
-
Invited From unknowns to insights: Bioinformatics for discovery in non-targeted screening.
International Conference on Non-Target Screening, Erding, Germany (October 13, 2025)
-
Invited A living benchmark to advance AI in proteomics de novo peptide sequencing.
Westlake Symposium for AI Proteomics and Virtual Cell, Hangzhou, China (October 9, 2025)
-
Poster A living proteomics benchmark for comprehensive evaluation of deep learning-based de novo peptide sequencing tools.
ASMS 2025, Baltimore, MD, USA (June 3, 2025)
-
Workshop Quality control for proteomics AI.
HUPO-PSI Spring Meeting 2025, Tübingen, Germany (March 31, 2025)
-
Invited Learning from repository-scale untargeted metabolomics data.
Metabolomics Society Early Career Members Network Webinar (January 22, 2025)
2024
-
Invited Artificial intelligence for molecular discovery to illuminate the dark proteome.
AOHUPO Online Educational Series (November 22, 2024)
-
Invited Sequence-to-sequence translation from mass spectra to peptides with a transformer model.
FoG Live (November 21, 2024)
-
Workshop Machine learning and AI in proteomics.
HUPO 2024, Dresden, Germany (October 21, 2024)
-
Invited Verborgen werelden ontrafeld: De rol van AI in het ontdekken van nieuwe biomedische geheimen.
Spectrum, Antwerp, Belgium (October 10, 2024)
-
Oral Sequence-to-sequence translation from mass spectra to peptides with a transformer model.
EuBIC-MS Seminar on Computational Mass Spectrometry, Barcelona, Spain (September 26, 2024)
-
Invited Learning from repository-scale untargeted metabolomics data.
Benelux Metabolomics Days, Ghent, Belgium (September 6, 2024)
-
Invited Learning from repository-scale untargeted metabolomics data.
UCR Post-ASMS Symposium, Riverside, CA, USA (June 7, 2024)
-
Workshop Machine learning for proteomics: Challenges, opportunities, and ProteomicsML.
ASMS 2024, Anaheim, CA, USA (June 5, 2024)
-
Workshop De novo peptide sequencing: Breakthroughs and challenges.
ASMS 2024, Anaheim, CA, USA (June 4, 2024)
-
Poster Enhanced spectrum clustering for molecular networking.
ASMS 2024, Anaheim, CA, USA (June 3, 2024)
-
Invited Improved immunopeptidome analysis using timsTOF fragment ion intensity prediction.
OMICS Research Day, Antwerp, Belgium (May 15, 2024)
-
Invited Molecules of mystery: A journey of computational mass spectrometry and artificial intelligence.
Faculty of Science Research Day, Antwerp, Belgium (February 8, 2024)
2023
-
Invited Bioinformatics to explore the molecular universe.
EuPA ECR Day (October 5, 2023)
-
Invited Sequence-to-sequence translation from mass spectra to peptides with a transformer model.
Proteomics ONLINE (September 28, 2023)
-
Workshop Machine learning applications for immunopeptidomics analysis.
Bioinformatics Hub, HUPO 2023 World Congress, Busan, South Korea (September 18, 2023)
-
Invited Deep learning to uncover the immunopeptidome.
BMSS 2023, Manchester, United Kingdom (September 14, 2023)
-
Oral Improved immunopeptidome analysis using timsTOF fragment ion intensity prediction.
BSPR/EuPA 2023, Newcastle upon Tyne, United Kingdom (July 19, 2023)
-
Invited Leveraging public mass spectrometry for molecular discovery from millions of mass spectra.
Biomina Lunch Talks, Antwerp, Belgium (June 30, 2023)
-
Poster Unified and standardized mass spectrometry data processing in Python using spectrum_utils.
ASMS 2023, Houston, TX, USA (June 5, 2023)
2022
-
Oral A learned embedding for efficient joint analysis of millions of mass spectra.
HUPO 2022 World Congress, Cancun, Mexico (December 7, 2022)
-
Poster Semi-supervised rescoring for open modification searching reveals protein modifications during SARS-CoV-2 infection.
HUPO 2022 World Congress, Cancun, Mexico (December 6, 2022)
-
Poster Comparison of spectrum similarity measures for discovery of structurally related molecules.
ASMS 2022, Minneapolis, MN, USA (June 8, 2022)
-
Invited A learned embedding for efficient joint analysis of millions of mass spectra.
Best of US HUPO 2022 (May 3, 2022)
-
Invited Leveraging public untargeted metabolomics data to propagate structurally related molecule annotations to millions of MS/MS spectra.
VIB Applied Bioinformatics in Life Sciences, Leuven, Belgium (March 11, 2022)
-
Oral A learned embedding for efficient joint analysis of millions of mass spectra.
US HUPO 2022, Charleston, SC, USA (March 1, 2022)
-
Poster Leveraging public untargeted metabolomics data to propagate annotations to millions of MS/MS spectra.
US HUPO 2022, Charleston, SC, USA (March 1, 2022)
2021
-
Workshop Quality control for proteomics and metabolomics.
ASMS 2021, Philadelphia, PA, USA (November 2, 2021)
-
Poster Repository scale propagated spectral library of suspects for untargeted metabolomics.
ASMS 2021, Philadelphia, PA, USA (November 2, 2021)
2020
-
Oral Repository-scale propagated spectral library of suspects.
SoCal Science @Home Lollapalooza (December 1, 2020)
-
Oral Deep neural network embedding for efficient repository-scale analysis of hundreds of millions of mass spectra.
ASMS 2020 Reboot (June 8, 2020)
-
Oral Identifying millions of protein modifications using GPU-powered approximate nearest neighbor searching.
GPU Technology Conference (March 26, 2020)
2019
-
Poster A learned embedding for efficient joint analysis of millions of mass spectra.
Machine Learning in Computational Biology, Vancouver, BC, Canada (December 14, 2019)
-
Poster ANN-SoLo: Extremely fast and accurate open modification spectral library searching.
ASMS 2019, Atlanta, GA, USA (June 3, 2019)
2018
-
Poster Fast open modification spectral library searching through approximate nearest neighbor indexing.
Department of Genome Sciences retreat, Leavenworth, WA, USA (September 17, 2018)
-
Oral Fast open modification spectral library searching through approximate nearest neighbor indexing.
Cascadia Proteomics Symposium, Seattle, WA, USA (July 23, 2018)
2017
-
Poster Mass spectrometry proteomics: Ready for the deep learning (r)evolution?
ASMS 2017, Indianapolis, IN, USA (June 8, 2017)
-
Poster Towards the smart lab: A comprehensive approach to mass spectrometry quality control.
ASMS 2017, Indianapolis, IN, USA (June 8, 2017)
-
Invited Shedding light on complex mass spectrometry proteomics processes through advanced data mining.
Faculty of Science Research Day, Antwerp, Belgium (January 13, 2017)
2016
-
Oral Git: How version control can power your research.
Biomina Lunch Talks, Antwerp, Belgium (June 24, 2016)
-
Oral Optimized open modification spectral library searching using approximate nearest neighbor techniques.
ASMS 2016, San Antonio, TX, USA (June 9, 2016)
-
Poster Automatic quality assessment of mass spectrometry experiments by multivariate quality control metrics.
ASMS 2016, San Antonio, TX, USA (June 9, 2016)
2015
-
Oral Approaches for mass spectrometry quality control.
Biomina Research Day, Antwerp, Belgium (December 9, 2015)
-
Oral Analysis of mass spectrometry quality control metrics.
Benelux Bioinformatics Conference, Antwerp, Belgium (December 7, 2015)
Outstanding oral presentation award
-
Oral Precursor-free and fast spectral library search using approximate nearest neighbor techniques.
Benelux Bioinformatics Conference Student Council Symposium, Antwerp, Belgium (December 6, 2015)
-
Poster Mass spectrometry quality control: Instrument monitoring and pattern mining insights.
ASMS 2015, St. Louis, MO, USA (June 2, 2015)
-
Workshop iMonDB: Mass spectrometry instrument monitoring for quality control.
Methods and tools for intra- and inter-experiment LC MS performance tracking workshop, ASMS 2015, St. Louis, MO, USA (June 1, 2015)
2014
-
Poster A comprehensive approach to mass spectrometry quality control.
Two-day symposium of the Belgian Proteomics Association, Brussels, Belgium (December 18, 2014)
-
Oral Pattern mining of mass spectrometry quality control data.
Benelux Bioinformatics Conference, Luxembourg, Luxembourg (December 9, 2014)
-
Poster Pattern mining of mass spectrometry quality control data.
European Conference on Computational Biology, Strasbourg, France (September 7, 2014)
-
Poster Mining mass spectrometry quality control data.
ASMS 2014, Baltimore, MD, USA (June 16, 2014)
-
Oral Collecting and mining mass spectrometry quality control data.
Biomina Research Day, Antwerp, Belgium (February 20, 2014)
2013
Selected software
See also GitHub.

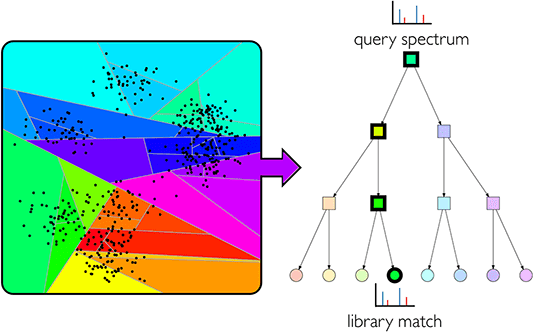
- GLEAMS
- Siamese deep neural network to embed mass spectra into a latent space.
- ANN-SoLo
- Fast and sensitive GPU-powered open modification spectral library searching.

- falcon
- Large-scale tandem mass spectrum clustering using fast nearest neighbor searching.
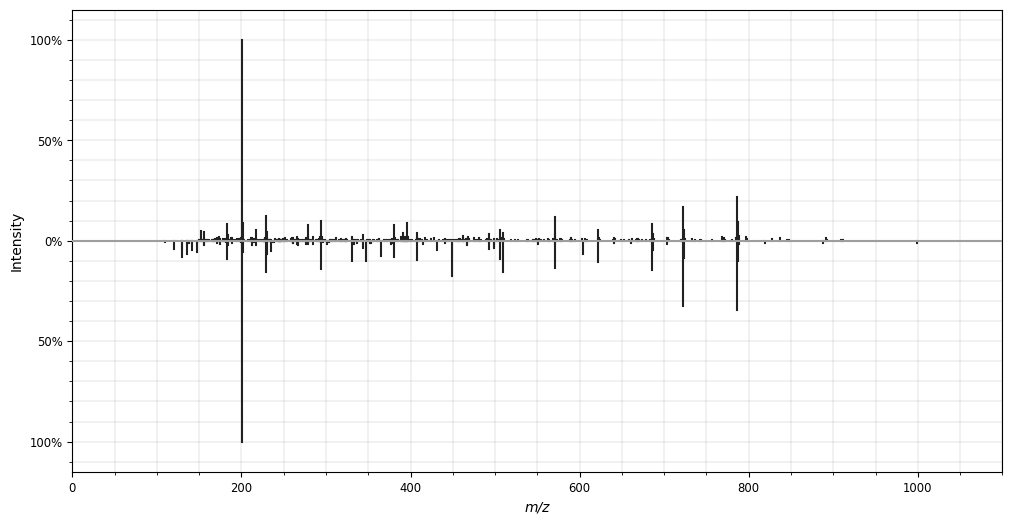
- spectrum_utils
- Python package for efficient mass spectrum processing and visualization.